Background
This is really a short post (note) for myself and probably for others who may be interested in software tools to visualise in silico macromolecules and small molecules.
Most bioinformaticians or structural biologists are probably already familiar with this software package, Molstar or Mol* (Sehnal et al. 2021). Molstar is a 3D viewer for large macromolecules (e.g. proteins), which are commonly used in structural biology and drug discovery (and also other related scientific disciplines).
A Quarto extension has been developed to embed the Molstar interactive 3D viewer inside Quarto markdown documents, which can be rendered as HTML pages. The main advantage of this is that it’s useful for reports or presentations.
Some useful links
Molstar webpage: https://molstar.org/
GitHub repository for the Quarto extension (thanks to the contributing team for this extension!): https://github.com/jmbuhr/quarto-molstar - example provided https://jmbuhr.de/quarto-molstar/
Other Molstar example: https://ljmartin.github.io/sideprojects/dockviz2.html
R Shiny and Molstar example (for people who prefer using R and R Shiny): https://www.appsilon.com/post/shiny-molstar-r-package-molecular-structures-visualizations
Streamlit and Dash integrations are also possible, this also makes me think that I could probably try integrating Molstar with Shiny for Python, it’ll likely be a future side project.
An example using Molstar with RCSB PDB
The following example retrieves a protein (PDB ID: 4MQT) from RCSB PDB.
{{< mol-rcsb 4mqt >}}
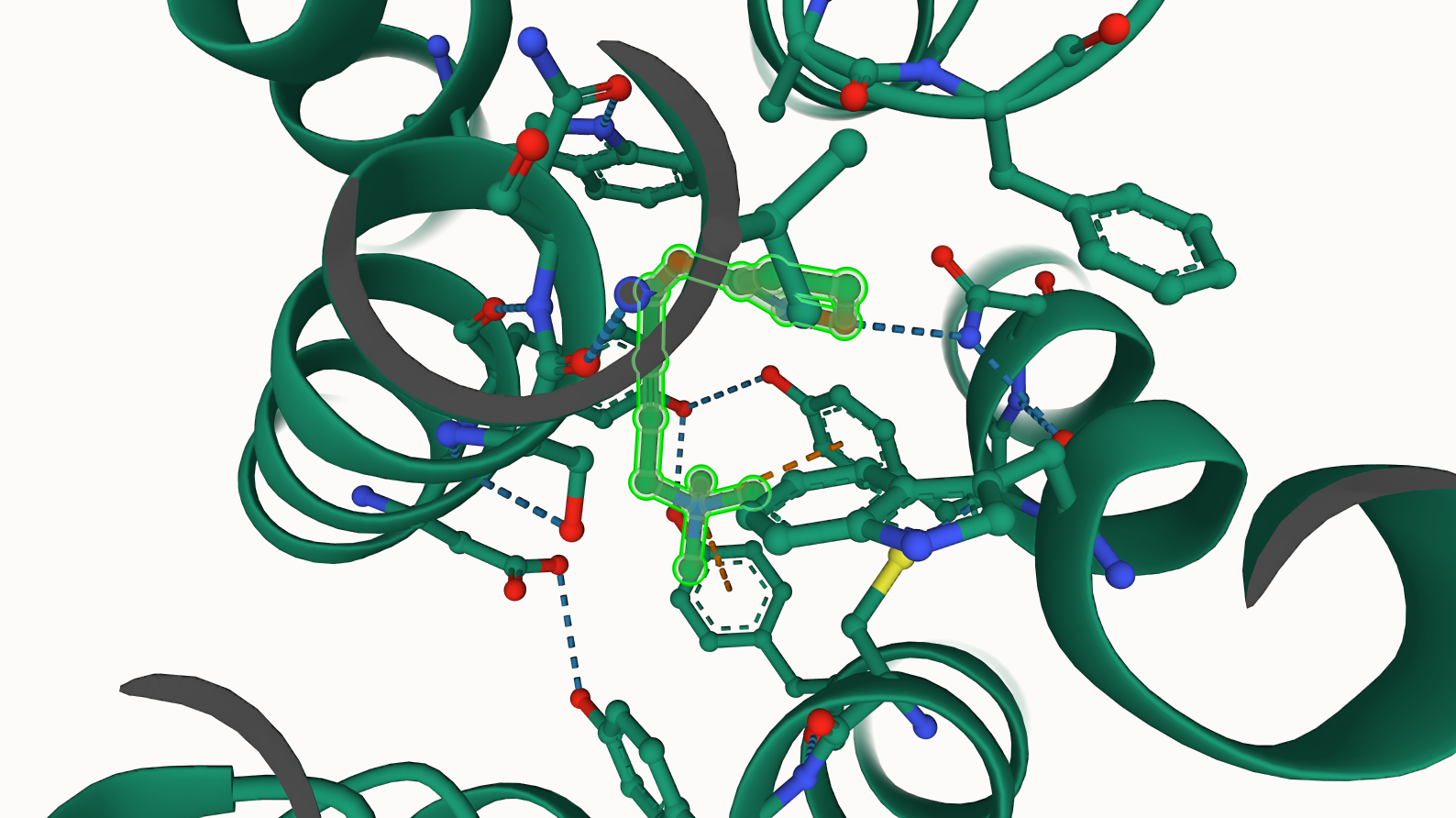
Hover over protein structure to see details of amino acid residues or ligands present in the structure.
To focus or zoom-in on the ligand bound to the receptor, just click on the ligand first. This shows most of the chemical interactions between the receptor and ligand bound to it (e.g. hydrogen bondings, other chemical interactions will appear if present e.g. pi-pi stacking).
Screenshots or state snapshots are also available from the viewer (other utility functions can be found on the top right corner of the viewer).
MD trajectories are also available, although I haven’t quite got there yet but it’s useful to know this may be possible (see example C from https://molstar.org/viewer-docs/examples/).
It’s also possible to upload AlphaFold-sourced proteins, or from other file sources (see examples shown from Molstar example).